The focus of our research is drug discovery using a variety of theoretical and experimental approaches. We are interested in modelling the structures of therapeutically important proteins particularly the cell surface membrane proteins. Recently, we have started to use the phage display technique as a powerful tool for drug design and drug delivery.
Molecular Modelling
Integral-membrane proteins (IMPs) accomplish a variety of important cellular functions and make up a large fraction of all proteins (Goffeau et al. 1993; Jones 1998) reflecting the fact that they play critical roles in maintaining the homeostasis and responsiveness of cells, organs, and organisms. Although like all other proteins, the information necessary for the folding of IMPs are embedded in their sequences, however, the folding pathways may seem rather specific (Booth & Curran 1999). Recently we have developed a new amino acid membrane propensity scale based on the structural analyses of non-redundant set of soluble proteins. The propensities were used successfully to predict the rotational orientation angle of membrane segments of the test IMPs (Dastmalchi et al. 2007).This method was implemented in a program called HTMSRAP which is available on line for public use . Please see Structural Bioinformatics and Molecular Design lab .

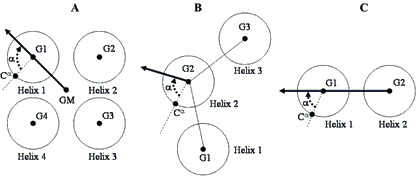 |
| Schematic representation of the rotational orientation angle, a, of the TM helices of IMP structures |
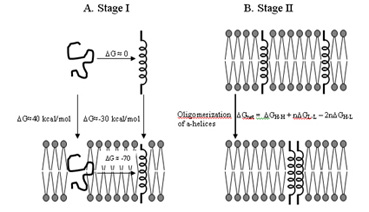 |
| Schematic representation of "two-stage" model for folding of IMPs (adapted from Lemmon et al. (1997) |
Application of Phage Display in Drug Development
Phage display technique is a powerful method to identify ligand(s) for a given target macromolecule. We are using peptide displaying phage library to identify peptide(s) which their absorption from GI may be facilitated via specific transporter molecules residing on the epithelial cells. Our preliminary results indicate that GI barrier shows no specificity toward the phage particles crossing this barrier. The sequences of the peptides displayed by the phages recovered from the blood or spleen of the rat after orally administration of peptide phage library do not demonstrate any consensus.
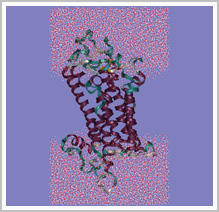 |
Carton representation of Histamine H3 receptor's structural model simulated in
water-vacuum-water environment |